miaverse:
R/Bioconductor framework for microbiome data science We are envisioning to rethink microbiome data science in R/Bioconductor. This is an open source project developing R/Bioc methods, benchmarking data, and educational material for microbiome research based on the SummarizedExperiment class and its derivatives. New contributions are very welcome.
Bioconductor
Bioconductor is open-source software for bioinformatics.
It includes currently over 2,000 R packages. See more from their website.

miaverse
miaverse is an actively developed R/Bioconductor framework for microbiome data science. It consists of multiple R/Bioc packages and tutorial book. The idea is not only to offer tools for microbiome downstream analysis but also to serve as a resource for valuable insights, offering guidance on conducting microbiome data analysis and developing effective microbiome data science workflows.
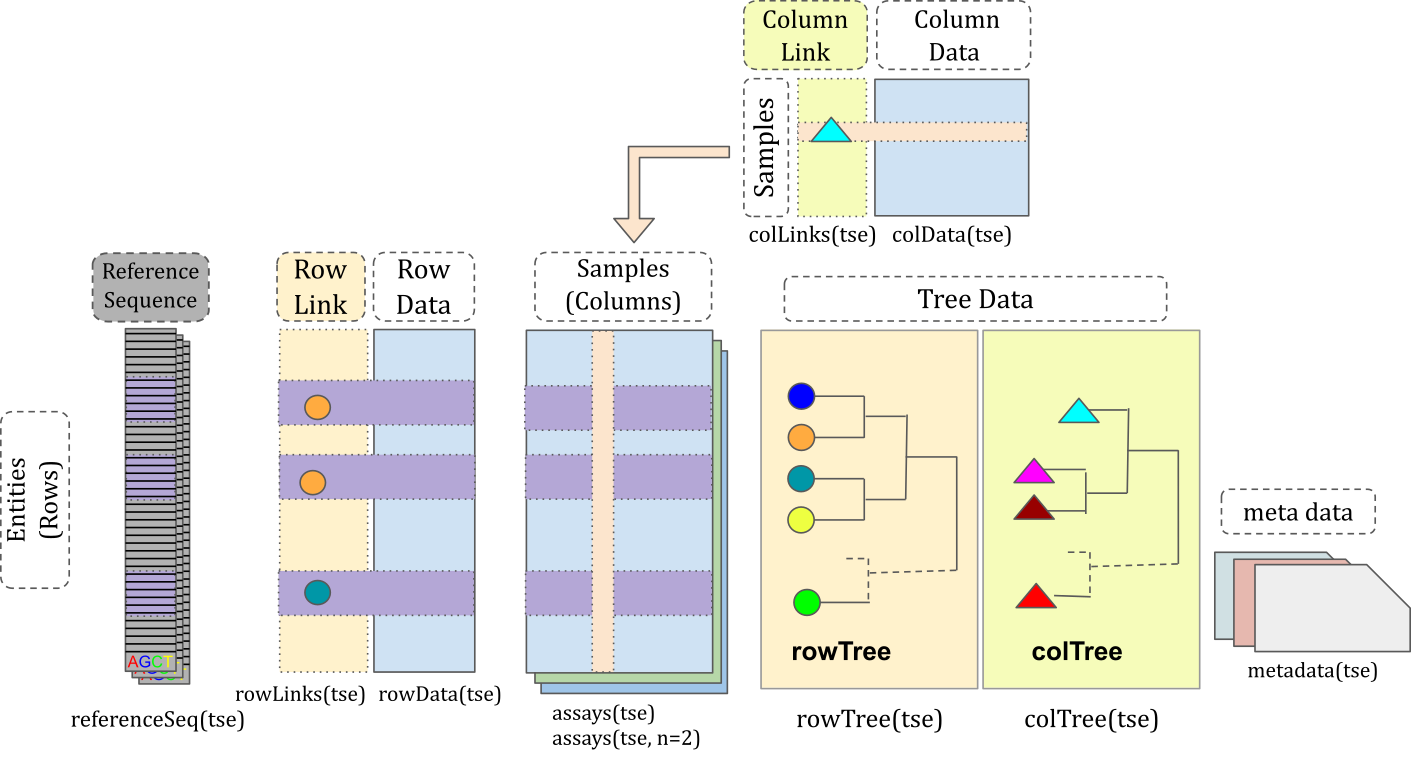
Data container
Within Bioconductor, software is rooted in the concept of data containers, which provide an
optimized framework for storing and managing data. This approach facilitates the efficient
development and utilization of methods.
miaverse specifically relies on the TreeSummarizedExperiment class, a tailored data container
optimized for microbiome data analysis.
Being built on the
SummarizedExperiment class, miaverse seamlessly integrates into the extensive SummarizedExperiment
ecosystem, ensuring a robust connection to a larger SummarizedExperiment ecosystem. This design choice
enhances the interoperability and versatility of miaverse in the broader Bioconductor context.
Methods
miaverse includes set of R/Bioc packages. Beyond the initial set of mia packages, the framework continues to evolve as an increasing number of method packages extend their support for miaverse. This expansion underscores the growing integration and compatibility of miaverse within the broader ecosystem, enriching its capabilities and ensuring users have access to a widening array of tools and methods for comprehensive microbiome data analysis.
Data
miaverse provides access to open data resources, allowing you to retrieve high-quality microbiome data for the development of methods and workflows.
See more from here.Documentation
The comprehensive tutorial book, Orchestrating Microbiome Analysis (OMA), not only demonstrates how to conduct microbiome data analysis but also includes example workflows for practical implementation. Below you can find relevant material.
Training
miaverse team has conducted numerous international courses and summer schools, relying on the OMA book as the primary training resource. OMA not only contains essential information but also includes practical exercises to support learning (link to exercies). Additionally, supplementary material is available; please see the Documentaton section.
Developers
The full list of contributors varies over time, and is identified through the individual repositories. Check each individual package for acknowledgements, contact information and references. Don't hesitate to contact us for feedback, support, feature requests, or if you want to contribute.